Produces a fit as per model_type plot with a facettable exposures/quantiles/distributions in ggplot2
Usage
ggresponseexpdist(
data = dplyr::filter(logistic_data, !is.na(ICGI)),
response = "response",
endpoint = "Endpoint",
model_type = c("loess", "linear", "logistic", "none"),
DOSE = "DOSE",
color_fill = "DOSE",
fit_by_color_fill = FALSE,
exposure_metrics = c("AUC", "CMAX"),
exposure_metric_split = c("median", "tertile", "quartile", "none"),
exposure_metric_soc_value = -99,
exposure_metric_plac_value = 0,
exposure_metric_soc_name = "SOC",
exposure_metric_plac_name = "Placebo",
exposure_distribution = c("distributions", "lineranges", "boxplots", "none"),
exposure_distribution_percent = c("none", "%", "N (%)", "N"),
exposure_distribution_Ntotal = c("none", "left", "right"),
exposure_distribution_percent_text_size = 5,
dose_plac_value = "Placebo",
xlab = "Exposure Values",
ylab = "Response",
points_alpha = 0.2,
points_show = TRUE,
mean_obs_byexptile = TRUE,
mean_obs_byexptile_plac = TRUE,
mean_obs_byexptile_text_size = 5,
mean_obs_byexptile_group = "none",
mean_obs_bydose = FALSE,
mean_obs_bydose_plac = FALSE,
mean_obs_bydose_text_size = 5,
N_byexptile_ypos = c("with means", "top", "bottom", "none"),
N_bydose_ypos = c("with means", "top", "bottom", "none"),
N_text_size = 5,
N_text_sep = NULL,
binlimits_show = TRUE,
binlimits_text_size = 5,
binlimits_ypos = 0,
binlimits_color = "#B3B3B380",
dist_position_scaler = 0.2,
dist_offset = 0,
dist_scale = 0.9,
lineranges_ypos = NULL,
lineranges_dodge = NULL,
lineranges_doselabel = FALSE,
lineranges_Ntotal = c("none", "left", "right"),
proj_bydose = FALSE,
yproj = FALSE,
yproj_xpos = 0,
yproj_dodge = 0.2,
yaxis_position = c("left", "right"),
facet_formula = NULL,
theme_certara = TRUE,
color_legend_title = "",
fill_legend_title = "",
linetype_legend_title = "",
shape_legend_title = "",
combine_fill_linetype_legend = TRUE,
legend_order = c("model", "color", "linetype", "shape"),
return_list = FALSE
)Arguments
- data
Data to use with multiple endpoints stacked into response (values), Endpoint(endpoint name)
- response
name of the column holding the response values
- endpoint
name of the column holding the name/key of the endpoint default to
Endpoint- model_type
type of the trend fit one of "loess", "linear", "logistic", "none"
- DOSE
name of the column holding the DOSE/regimen values default to
DOSEshould be a factor- color_fill
name of the column to be used for color/fill default to DOSE column should be a factor
- fit_by_color_fill
split fit by color/fill? default
FALSE- exposure_metrics
name(s) of the column(s) to be stacked into
expnameexptileand split intoexposure_metric_split- exposure_metric_split
one of "median", "tertile", "quartile", "none"
- exposure_metric_soc_value
special exposure code for standard of care default -99
- exposure_metric_plac_value
special exposure code for placebo default 0
- exposure_metric_soc_name
soc name default to "soc"
- exposure_metric_plac_name
placebo name default to "placebo"
- exposure_distribution
one of distributions, lineranges, boxplots or none
- exposure_distribution_percent
show N/percent of distribution between binlimits one of "%", "N (%)","N","none"
- exposure_distribution_Ntotal
show Ntotal by dose level next to the distribution one of "left","right","none"
- exposure_distribution_percent_text_size
distribution percentages text size default to 5
- dose_plac_value
string identifying placebo in DOSE column
- xlab
text to be used as x axis label
- ylab
text to be used as y axis label
- points_alpha
alpha transparency for points
- points_show
show the observations
TRUE/FALSE- mean_obs_byexptile
observed mean by exptile
TRUE/FALSE- mean_obs_byexptile_plac
observed mean by exptile placebo
TRUE/FALSE- mean_obs_byexptile_text_size
by exptile mean text size default to 5
- mean_obs_byexptile_group
additional grouping for exptile means default
none- mean_obs_bydose
observed mean by dose
TRUE/FALSE- mean_obs_bydose_plac
observed mean by placebo dose
TRUE/FALSE- mean_obs_bydose_text_size
by dose mean text size default to 5
- N_byexptile_ypos
N responders/Ntotal y position by exptile one of
with meanstopbottomnone- N_bydose_ypos
N responders/Ntotal y position by dose/color one of
with meanstopbottomnone- N_text_size
N responders/Ntotal text size default to 5
- N_text_sep
character string to separate N responders/Ntotal or N/mean defaults to
/otherwise\n- binlimits_show
show the binlimits vertical lines
TRUE/FALSE- binlimits_text_size
binlimits text size default to 5
- binlimits_ypos
binlimits y position default to -Inf
- binlimits_color
binlimits text color default to alpha("gray70",0.5)
- dist_position_scaler
space occupied by the distribution default to 0.2
- dist_offset
offset where the distribution position starts default to 0
- dist_scale
scaling parameter for ggridges default to 0.9
- lineranges_ypos
where to put the lineranges -1
- lineranges_dodge
lineranges vertical dodge value 1
- lineranges_doselabel
TRUE/FALSE- lineranges_Ntotal
show Ntotal by dose level next to the lineranges one of "left","right","none"
- proj_bydose
project the predictions on the fit curve
TRUE/FALSE- yproj
project the predictions on y axis
TRUE/FALSE- yproj_xpos
y projection x position 0
- yproj_dodge
y projection dodge value 0.2
- yaxis_position
where to put y axis "left" or "right"
- facet_formula
facet formula to be use otherwise
endpoint ~ expname- theme_certara
apply certara colors and format for strips and default colour/fill
- color_legend_title
text for colour legend title
- fill_legend_title
text for fill legend title
- linetype_legend_title
text for linetype legend title
- shape_legend_title
text for shape legend title
- combine_fill_linetype_legend
defaults to true
- legend_order
Legend order. A four-element vector with the following items ordered in your desired order: "model", "color", "linetype", "shape". if an item is absent the legend will be omitted.
- return_list
What to return if True a list of the datasets and plot is returned instead of only the plot
Examples
# Example 1
library(ggplot2)
effICGI <- logistic_data |>
dplyr::filter(!is.na(ICGI))|>
dplyr::filter(!is.na(AUC))
effICGI$DOSE <- factor(effICGI$DOSE,
levels=c("0", "600", "1200","1800","2400"),
labels=c("Placebo", "600 mg", "1200 mg","1800 mg","2400 mg"))
effICGI$STUDY <- factor(effICGI$STUDY)
effICGI$ICGI2 <- effICGI$ICGI
effICGI <- tidyr::gather(effICGI,Endpoint,response,ICGI,ICGI2)
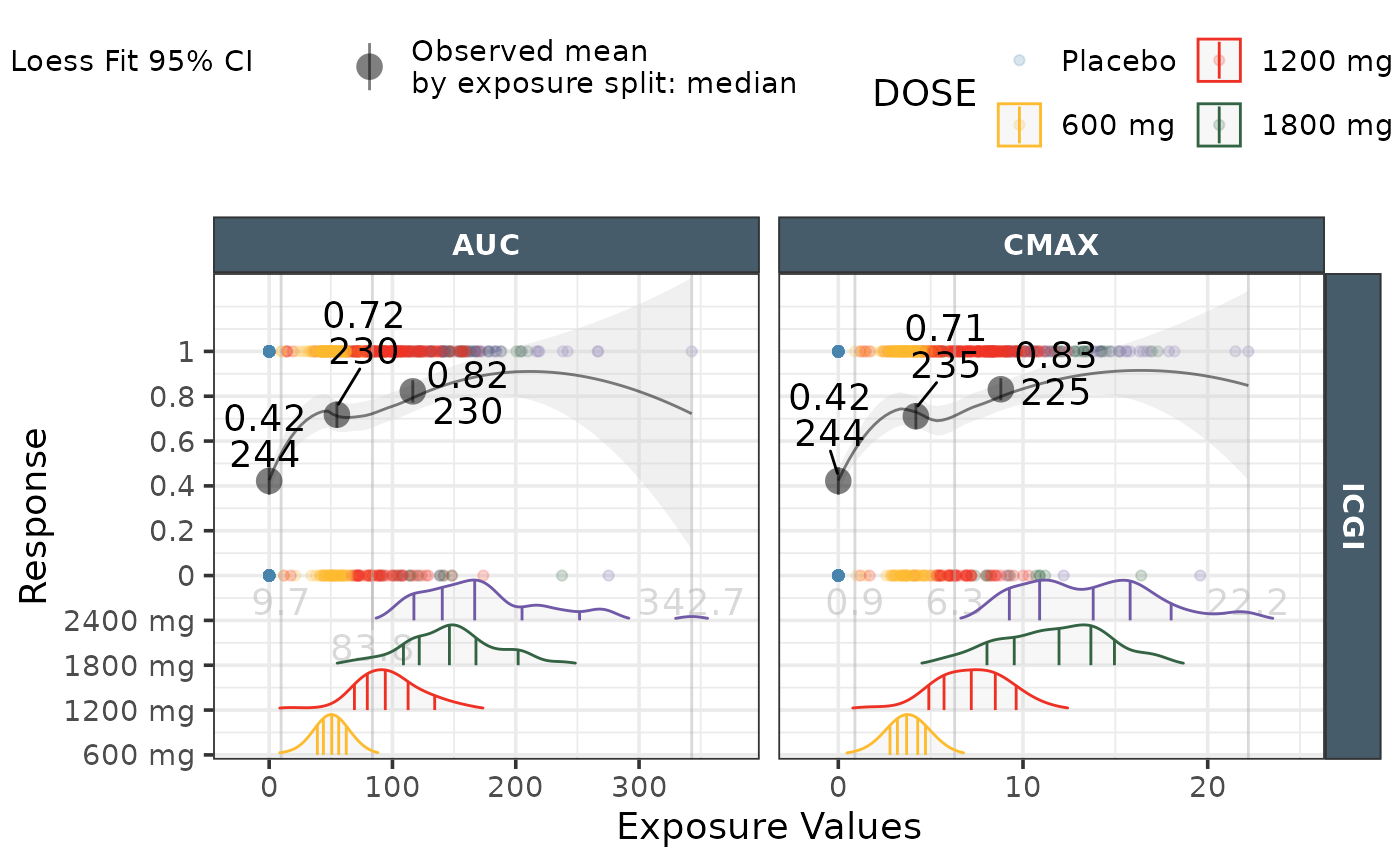
ggresponseexpdist(data = effICGI |>
dplyr::filter(Endpoint=="ICGI"),
model_type = "loess",
exposure_metrics = c("AUC","CMAX"),
legend_order = c("color","shape","model"),
color_legend_title ="Dose\nLevels")
#> Joining with `by = join_by(loopvariable, DOSE, quant_10)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_90)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_25)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_75)`
#> Joining with `by = join_by(loopvariable, DOSE, medexp)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_10)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_90)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_25)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_75)`
#> Joining with `by = join_by(loopvariable, DOSE, medexp)`
#> `geom_smooth()` using formula = 'y ~ x'
#> `geom_smooth()` using formula = 'y ~ x'
#> Picking joint bandwidth of 11.7
#> Picking joint bandwidth of 0.934
#> Warning: Removed 488 rows containing non-finite outside the scale range
#> (`stat_density_ridges()`).
 # Example 2
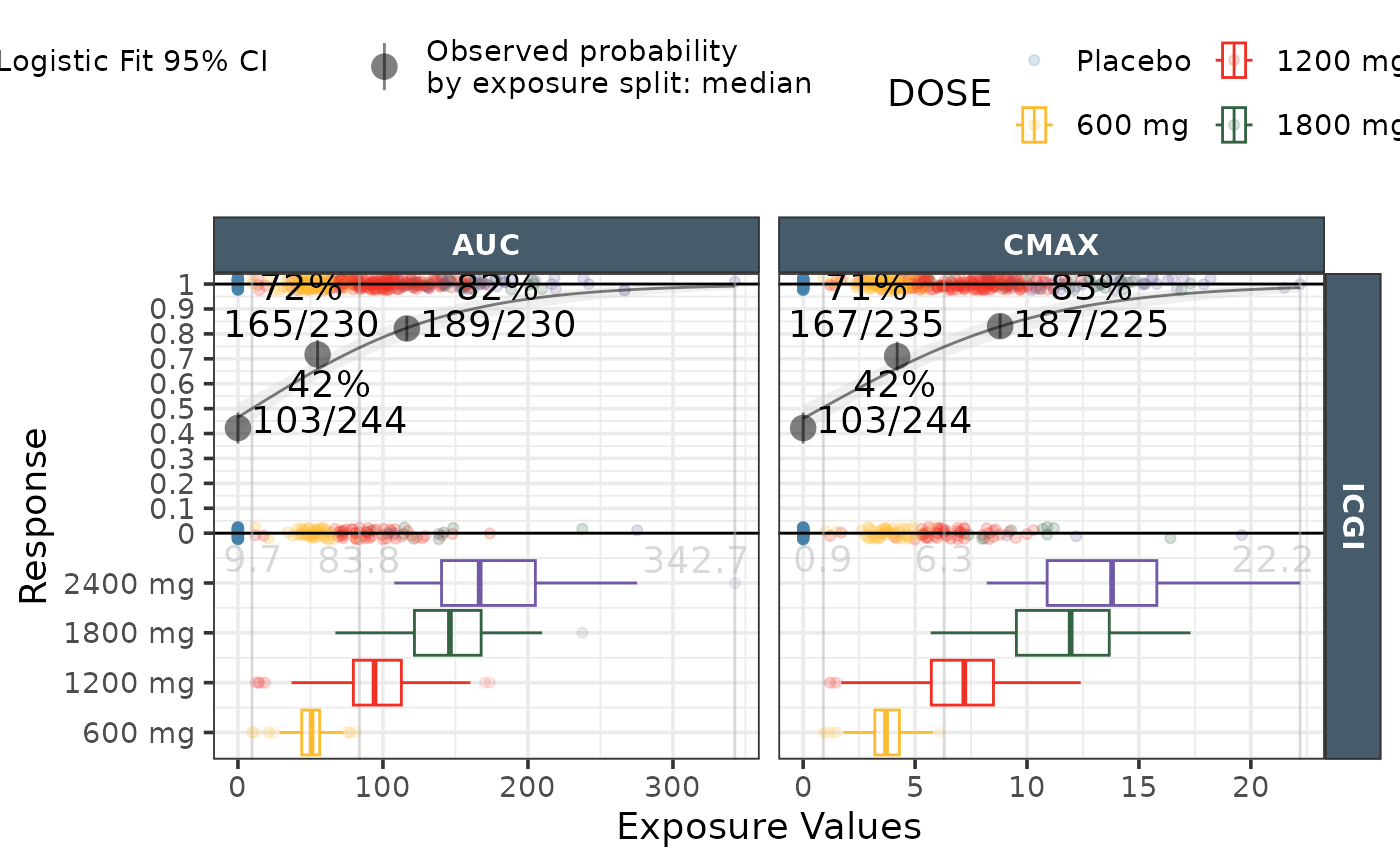
ggresponseexpdist(data = effICGI |>
dplyr::filter(Endpoint=="ICGI"),
model_type = "logistic",
exposure_metrics = c("AUC","CMAX"),
exposure_distribution ="boxplots")
#> Joining with `by = join_by(loopvariable, DOSE, quant_10)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_90)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_25)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_75)`
#> Joining with `by = join_by(loopvariable, DOSE, medexp)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_10)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_90)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_25)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_75)`
#> Joining with `by = join_by(loopvariable, DOSE, medexp)`
#> `geom_smooth()` using formula = 'y ~ x'
#> `geom_smooth()` using formula = 'y ~ x'
#> Warning: Removed 488 rows containing non-finite outside the scale range
#> (`stat_boxplot()`).
# Example 2
ggresponseexpdist(data = effICGI |>
dplyr::filter(Endpoint=="ICGI"),
model_type = "logistic",
exposure_metrics = c("AUC","CMAX"),
exposure_distribution ="boxplots")
#> Joining with `by = join_by(loopvariable, DOSE, quant_10)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_90)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_25)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_75)`
#> Joining with `by = join_by(loopvariable, DOSE, medexp)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_10)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_90)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_25)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_75)`
#> Joining with `by = join_by(loopvariable, DOSE, medexp)`
#> `geom_smooth()` using formula = 'y ~ x'
#> `geom_smooth()` using formula = 'y ~ x'
#> Warning: Removed 488 rows containing non-finite outside the scale range
#> (`stat_boxplot()`).
 # Example 3
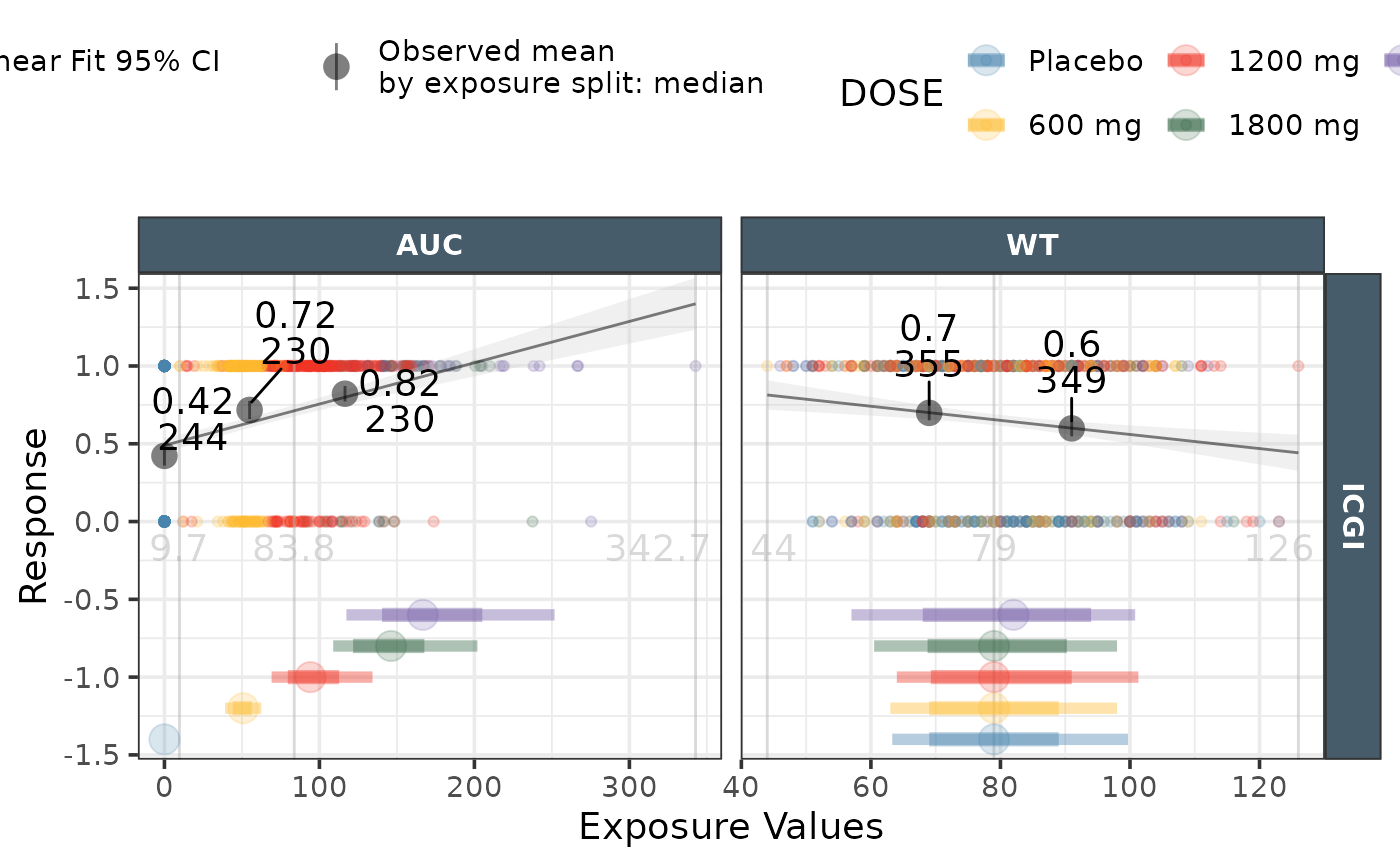
ggresponseexpdist(data = effICGI|>
dplyr::filter(Endpoint=="ICGI"),
model_type = "linear",
exposure_metrics = c("AUC","WT"),
exposure_distribution ="lineranges")
#> Joining with `by = join_by(loopvariable, DOSE, quant_10)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_90)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_25)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_75)`
#> Joining with `by = join_by(loopvariable, DOSE, medexp)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_10)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_90)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_25)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_75)`
#> Joining with `by = join_by(loopvariable, DOSE, medexp)`
#> `geom_smooth()` using formula = 'y ~ x'
#> Warning: In lm.wfit(x, y, w, offset = offset, singular.ok = singular.ok,
#> ...) :
#> extra argument ‘family’ will be disregarded
#> Warning: In lm.wfit(x, y, w, offset = offset, singular.ok = singular.ok,
#> ...) :
#> extra argument ‘family’ will be disregarded
#> `geom_smooth()` using formula = 'y ~ x'
#> Warning: In lm.wfit(x, y, w, offset = offset, singular.ok = singular.ok,
#> ...) :
#> extra argument ‘family’ will be disregarded
#> Warning: In lm.wfit(x, y, w, offset = offset, singular.ok = singular.ok,
#> ...) :
#> extra argument ‘family’ will be disregarded
# Example 3
ggresponseexpdist(data = effICGI|>
dplyr::filter(Endpoint=="ICGI"),
model_type = "linear",
exposure_metrics = c("AUC","WT"),
exposure_distribution ="lineranges")
#> Joining with `by = join_by(loopvariable, DOSE, quant_10)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_90)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_25)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_75)`
#> Joining with `by = join_by(loopvariable, DOSE, medexp)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_10)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_90)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_25)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_75)`
#> Joining with `by = join_by(loopvariable, DOSE, medexp)`
#> `geom_smooth()` using formula = 'y ~ x'
#> Warning: In lm.wfit(x, y, w, offset = offset, singular.ok = singular.ok,
#> ...) :
#> extra argument ‘family’ will be disregarded
#> Warning: In lm.wfit(x, y, w, offset = offset, singular.ok = singular.ok,
#> ...) :
#> extra argument ‘family’ will be disregarded
#> `geom_smooth()` using formula = 'y ~ x'
#> Warning: In lm.wfit(x, y, w, offset = offset, singular.ok = singular.ok,
#> ...) :
#> extra argument ‘family’ will be disregarded
#> Warning: In lm.wfit(x, y, w, offset = offset, singular.ok = singular.ok,
#> ...) :
#> extra argument ‘family’ will be disregarded
 # Example 4
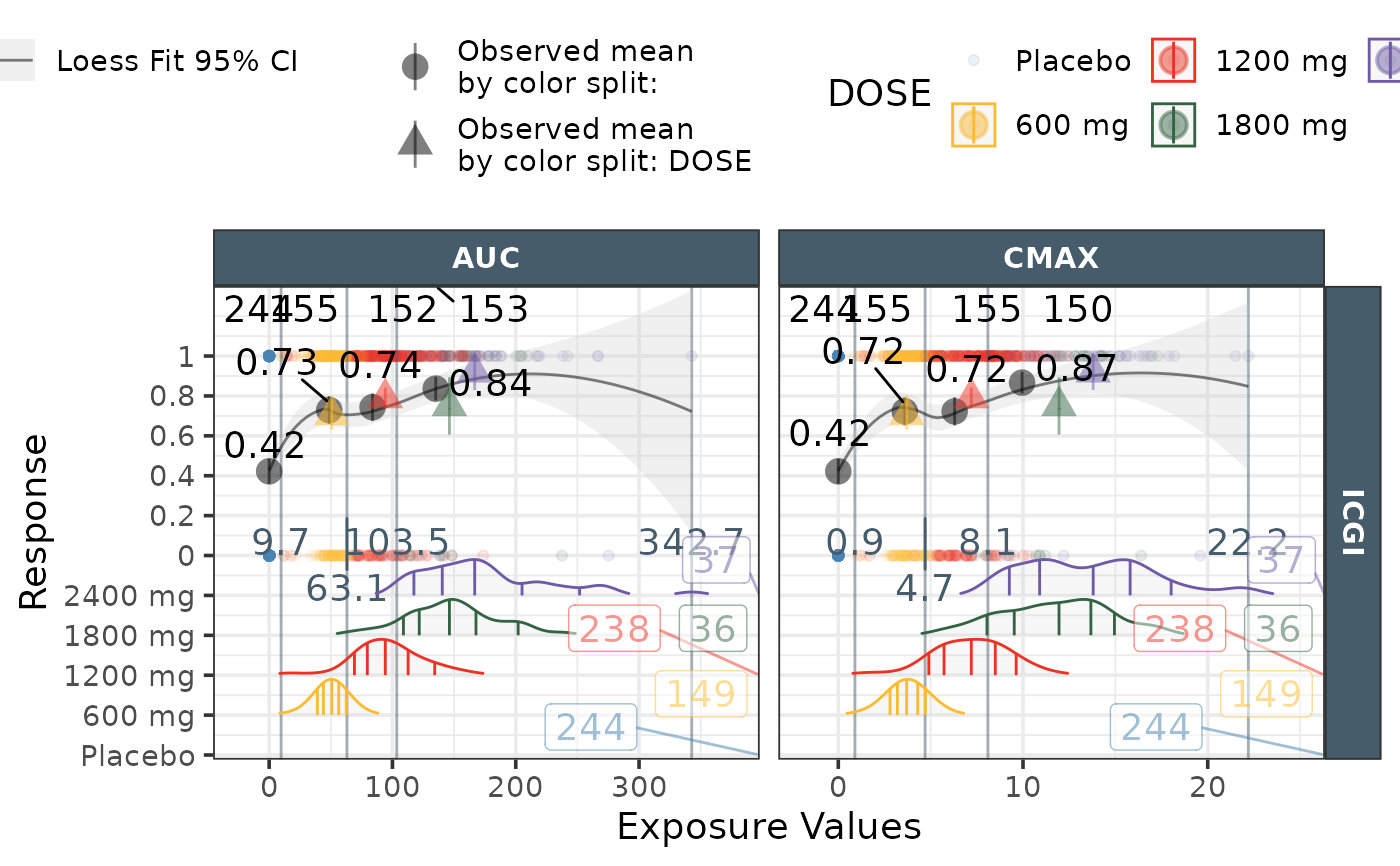
ggresponseexpdist(data = effICGI |>
dplyr::filter(Endpoint=="ICGI"),
response = "response",
endpoint = "Endpoint",
model_type = "loess",
DOSE = "DOSE",
color_fill = "DOSE",
exposure_metrics = c("AUC","CMAX"),
exposure_metric_split = c("tertile"),
exposure_metric_soc_value = -99,
exposure_metric_plac_value = 0,
exposure_distribution ="distributions",
exposure_distribution_Ntotal="right",
N_byexptile_ypos = "top",
mean_obs_bydose = TRUE,
mean_obs_bydose_text_size = 0,
mean_obs_byexptile_text_size = 5,
N_bydose_ypos = "none",
N_text_sep = "/",
binlimits_color = "#475c6b",
binlimits_ypos = 0.2,
points_alpha= 0.1)
#> Joining with `by = join_by(loopvariable, DOSE, quant_10)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_90)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_25)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_75)`
#> Joining with `by = join_by(loopvariable, DOSE, medexp)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_10)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_90)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_25)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_75)`
#> Joining with `by = join_by(loopvariable, DOSE, medexp)`
#> `geom_smooth()` using formula = 'y ~ x'
#> `geom_smooth()` using formula = 'y ~ x'
#> Picking joint bandwidth of 11.7
#> Picking joint bandwidth of 0.934
#> Warning: Removed 488 rows containing non-finite outside the scale range
#> (`stat_density_ridges()`).
# Example 4
ggresponseexpdist(data = effICGI |>
dplyr::filter(Endpoint=="ICGI"),
response = "response",
endpoint = "Endpoint",
model_type = "loess",
DOSE = "DOSE",
color_fill = "DOSE",
exposure_metrics = c("AUC","CMAX"),
exposure_metric_split = c("tertile"),
exposure_metric_soc_value = -99,
exposure_metric_plac_value = 0,
exposure_distribution ="distributions",
exposure_distribution_Ntotal="right",
N_byexptile_ypos = "top",
mean_obs_bydose = TRUE,
mean_obs_bydose_text_size = 0,
mean_obs_byexptile_text_size = 5,
N_bydose_ypos = "none",
N_text_sep = "/",
binlimits_color = "#475c6b",
binlimits_ypos = 0.2,
points_alpha= 0.1)
#> Joining with `by = join_by(loopvariable, DOSE, quant_10)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_90)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_25)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_75)`
#> Joining with `by = join_by(loopvariable, DOSE, medexp)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_10)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_90)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_25)`
#> Joining with `by = join_by(loopvariable, DOSE, quant_75)`
#> Joining with `by = join_by(loopvariable, DOSE, medexp)`
#> `geom_smooth()` using formula = 'y ~ x'
#> `geom_smooth()` using formula = 'y ~ x'
#> Picking joint bandwidth of 11.7
#> Picking joint bandwidth of 0.934
#> Warning: Removed 488 rows containing non-finite outside the scale range
#> (`stat_density_ridges()`).
 if (FALSE) { # \dontrun{
# Example 5
effICGI <- logistic_data |>
dplyr::filter(!is.na(ICGI))|>
dplyr::filter(!is.na(AUC))
effICGI$DOSE <- factor(effICGI$DOSE,
levels=c("0", "600", "1200","1800","2400"),
labels=c("Placebo", "600 mg", "1200 mg","1800 mg","2400 mg"))
effICGI$STUDY <- factor(effICGI$STUDY)
effICGI$ICGI2 <- ifelse(effICGI$ICGI7 < 4,1,0)
effICGI$ICGI3 <- ifelse(effICGI$ICGI7 < 5,1,0)
effICGI <- tidyr::gather(effICGI,Endpoint,response,ICGI,ICGI2,ICGI3)
effICGI$endpointcol2 <- effICGI$Endpoint
effICGI$endpointcol3 <- effICGI$Endpoint
ggresponseexpdist(data = effICGI,
points_show = FALSE,
exposure_metrics = c("AUC"),
exposure_distribution ="lineranges",
color_fill = "endpointcol2",
model_type = "logistic",
fit_by_color_fill = TRUE,
mean_obs_byexptile_text_size = 0,
mean_obs_byexptile_group="endpointcol3",
facet_formula = Endpoint~expname,
N_byexptile_ypos = "none",
binlimits_text_size = 0
)+
ggplot2::facet_grid(expname~Endpoint,margin="Endpoint")
# Example 6 retrun a list and customize
plist <- ggresponseexpdist(data = effICGI |>
dplyr::filter(Endpoint=="ICGI"),
model_type = "logistic",
mean_obs_byexptile = TRUE,
N_byexptile_ypos = "none",
mean_obs_byexptile_text_size = 4,
binlimits_ypos = -Inf,
exposure_metric_split = "tertile",
exposure_metrics = c("AUC"),
return_list = TRUE)
byexptileinformation <- plist[[7]]
plotwihoutlabels <- plist[[9]]
#construct text label
byexptileinformation <- byexptileinformation %>%
dplyr::group_by(exptile,expname)%>%
dplyr::mutate(meanbin = mean(c(minexp,maxexp))) %>%
dplyr::mutate(label = ifelse(exptile!="Placebo",
paste0(exptile,"\n","[",round(minexp,0),"-",round(maxexp,0),"]",
"\n",N,"\n",Ntot),
paste0(exptile,"\n","",
"\n",N,"\n",Ntot)),
x_pos = ifelse(exptile=="Placebo",-25,meanbin )
)
plotwihoutlabels +
geom_text(data=byexptileinformation,size = 3,
aes(x=x_pos ,label=label,y = 0.5),
inherit.aes = FALSE,
vjust= 1, hjust = 0.5)+
geom_text(data=data.frame(
label="exposure ntile\n[min-max]\nN responders\nN Total"),size = 3,
x=Inf,y = 0.5,
aes(label=label),inherit.aes = FALSE,
vjust= 1, hjust = 1)+
facet_wrap(~expname,ncol=2,scales = "free_x")
} # }
if (FALSE) { # \dontrun{
# Example 5
effICGI <- logistic_data |>
dplyr::filter(!is.na(ICGI))|>
dplyr::filter(!is.na(AUC))
effICGI$DOSE <- factor(effICGI$DOSE,
levels=c("0", "600", "1200","1800","2400"),
labels=c("Placebo", "600 mg", "1200 mg","1800 mg","2400 mg"))
effICGI$STUDY <- factor(effICGI$STUDY)
effICGI$ICGI2 <- ifelse(effICGI$ICGI7 < 4,1,0)
effICGI$ICGI3 <- ifelse(effICGI$ICGI7 < 5,1,0)
effICGI <- tidyr::gather(effICGI,Endpoint,response,ICGI,ICGI2,ICGI3)
effICGI$endpointcol2 <- effICGI$Endpoint
effICGI$endpointcol3 <- effICGI$Endpoint
ggresponseexpdist(data = effICGI,
points_show = FALSE,
exposure_metrics = c("AUC"),
exposure_distribution ="lineranges",
color_fill = "endpointcol2",
model_type = "logistic",
fit_by_color_fill = TRUE,
mean_obs_byexptile_text_size = 0,
mean_obs_byexptile_group="endpointcol3",
facet_formula = Endpoint~expname,
N_byexptile_ypos = "none",
binlimits_text_size = 0
)+
ggplot2::facet_grid(expname~Endpoint,margin="Endpoint")
# Example 6 retrun a list and customize
plist <- ggresponseexpdist(data = effICGI |>
dplyr::filter(Endpoint=="ICGI"),
model_type = "logistic",
mean_obs_byexptile = TRUE,
N_byexptile_ypos = "none",
mean_obs_byexptile_text_size = 4,
binlimits_ypos = -Inf,
exposure_metric_split = "tertile",
exposure_metrics = c("AUC"),
return_list = TRUE)
byexptileinformation <- plist[[7]]
plotwihoutlabels <- plist[[9]]
#construct text label
byexptileinformation <- byexptileinformation %>%
dplyr::group_by(exptile,expname)%>%
dplyr::mutate(meanbin = mean(c(minexp,maxexp))) %>%
dplyr::mutate(label = ifelse(exptile!="Placebo",
paste0(exptile,"\n","[",round(minexp,0),"-",round(maxexp,0),"]",
"\n",N,"\n",Ntot),
paste0(exptile,"\n","",
"\n",N,"\n",Ntot)),
x_pos = ifelse(exptile=="Placebo",-25,meanbin )
)
plotwihoutlabels +
geom_text(data=byexptileinformation,size = 3,
aes(x=x_pos ,label=label,y = 0.5),
inherit.aes = FALSE,
vjust= 1, hjust = 0.5)+
geom_text(data=data.frame(
label="exposure ntile\n[min-max]\nN responders\nN Total"),size = 3,
x=Inf,y = 0.5,
aes(label=label),inherit.aes = FALSE,
vjust= 1, hjust = 1)+
facet_wrap(~expname,ncol=2,scales = "free_x")
} # }
