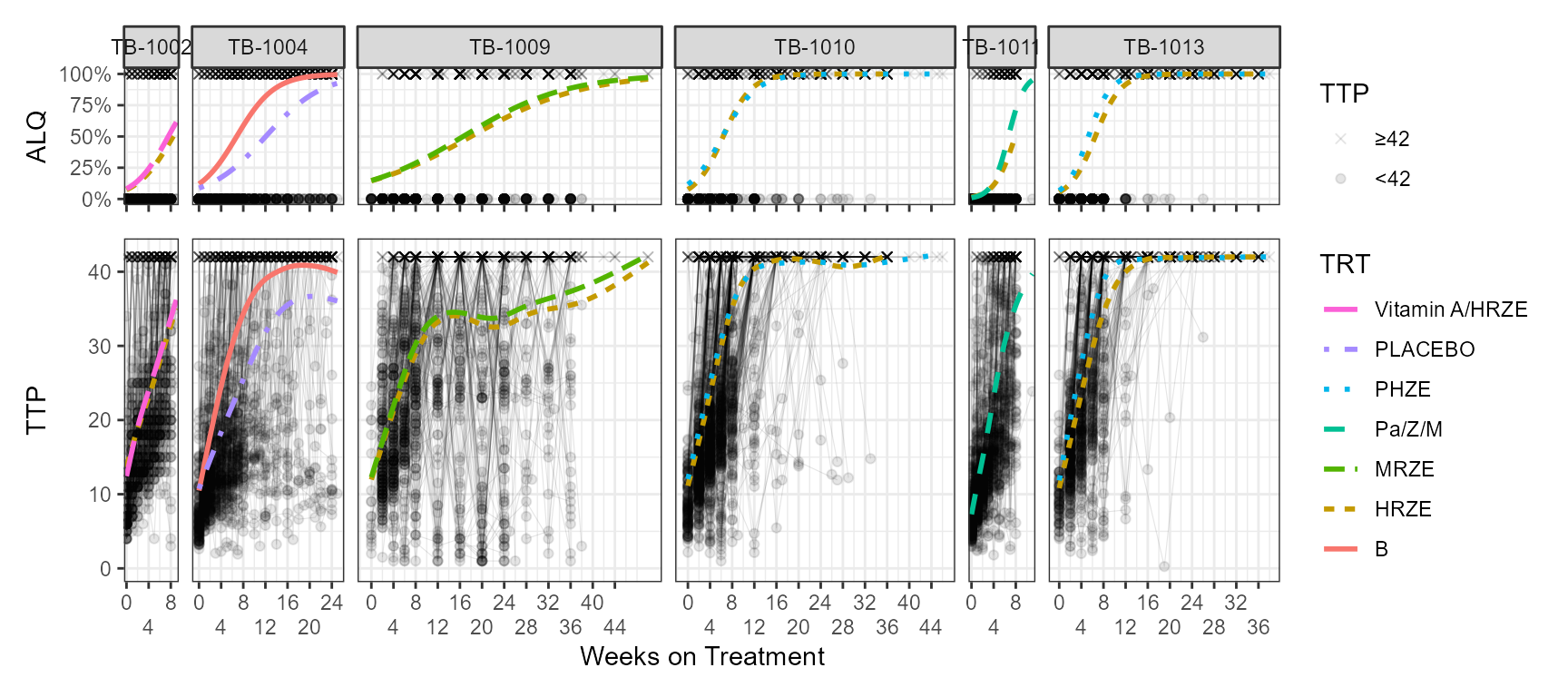
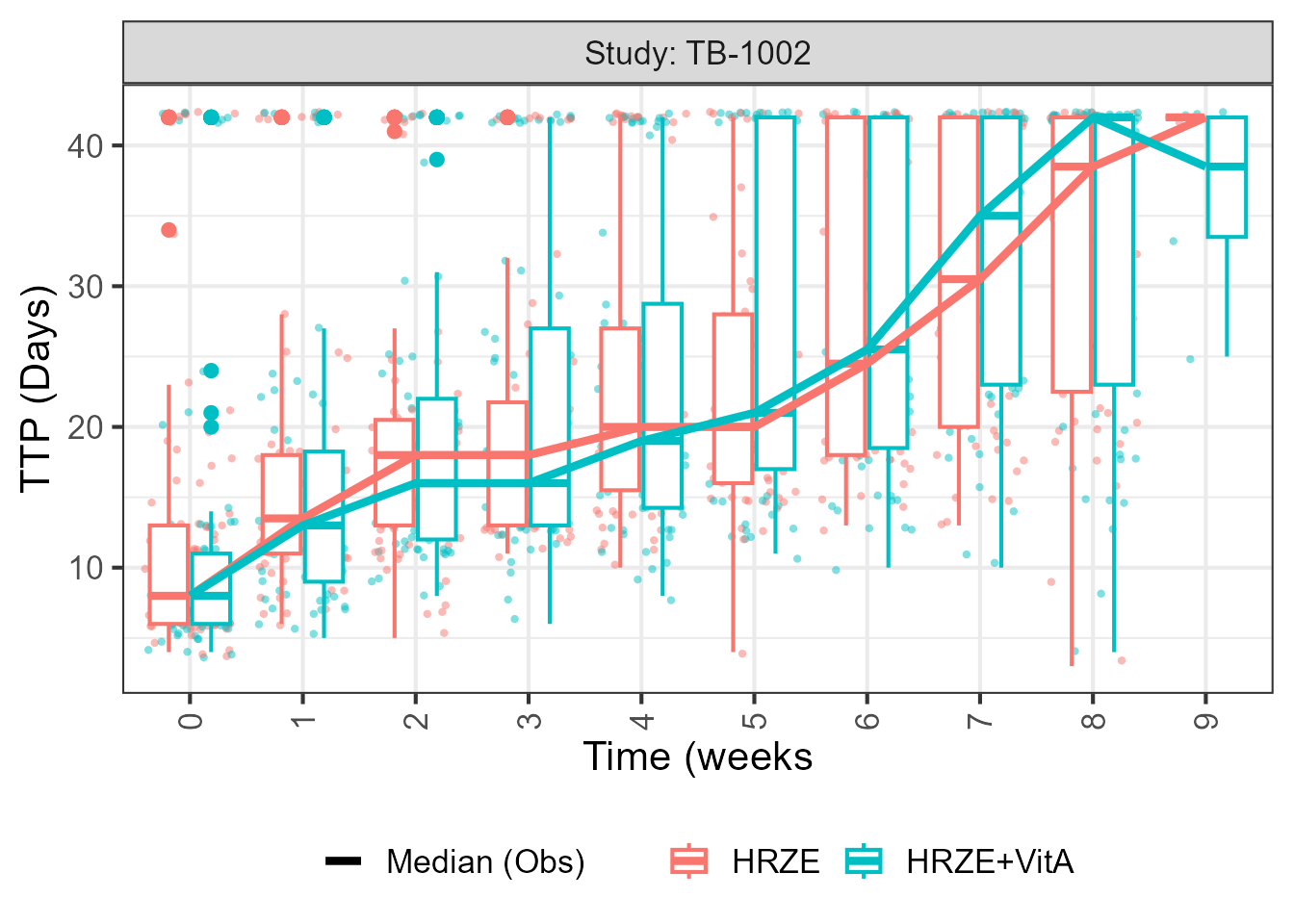
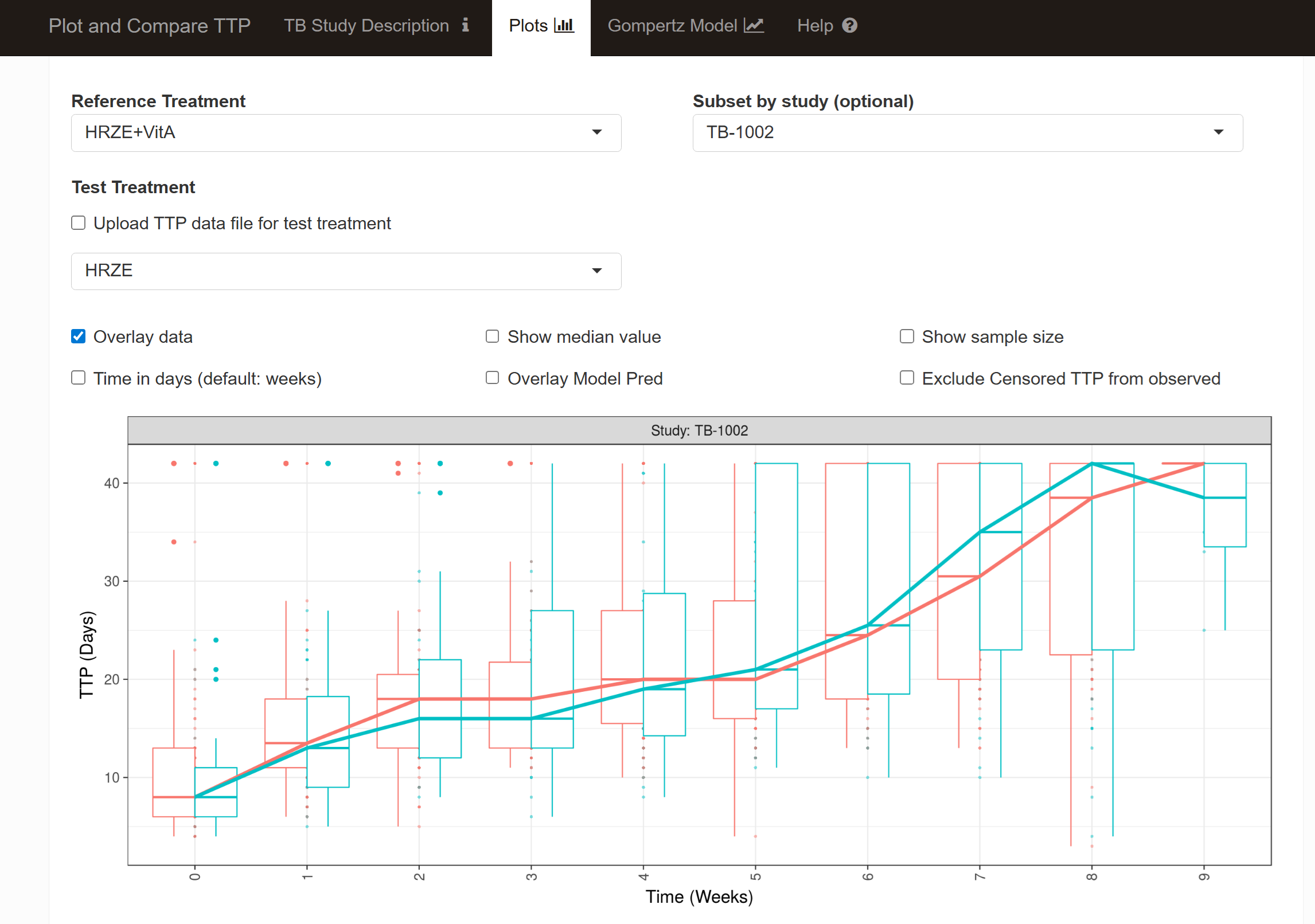
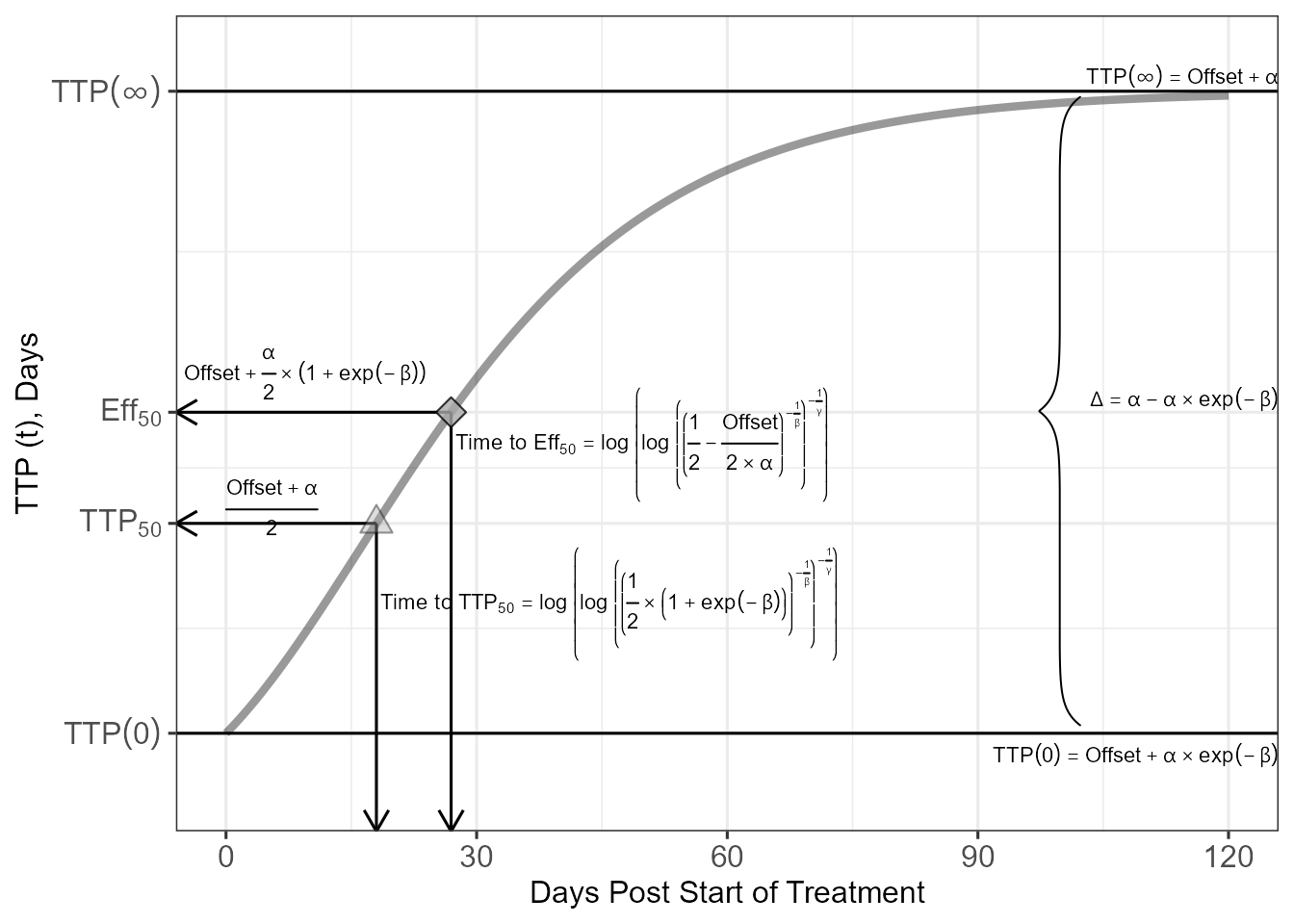
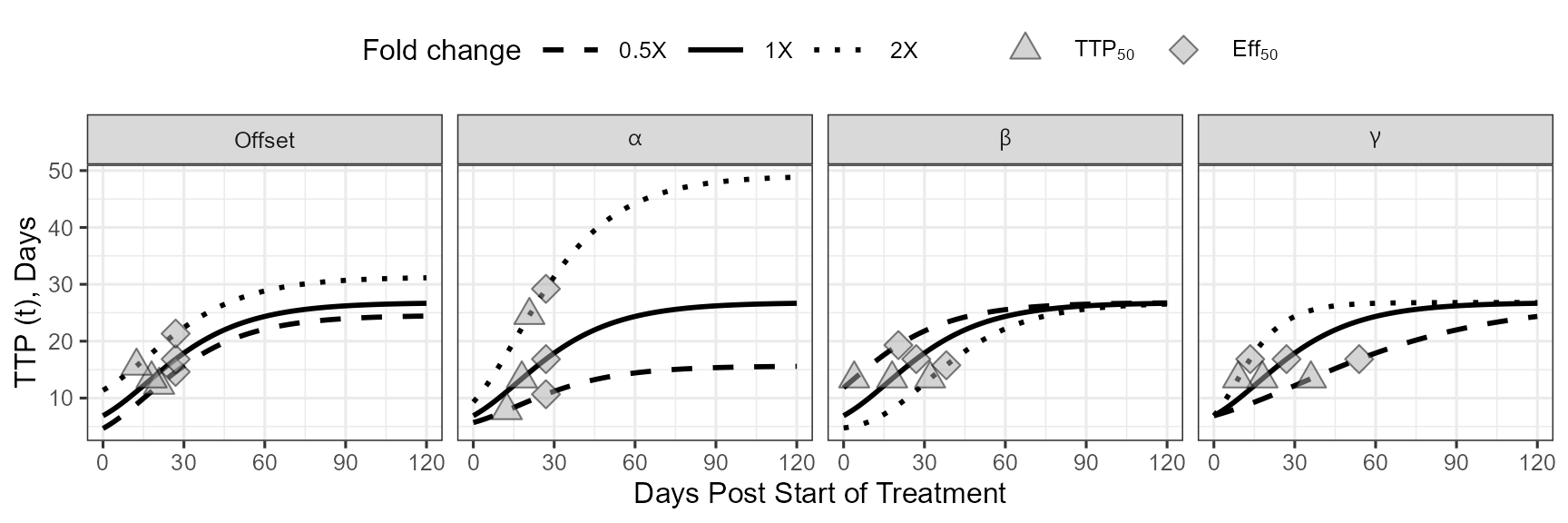
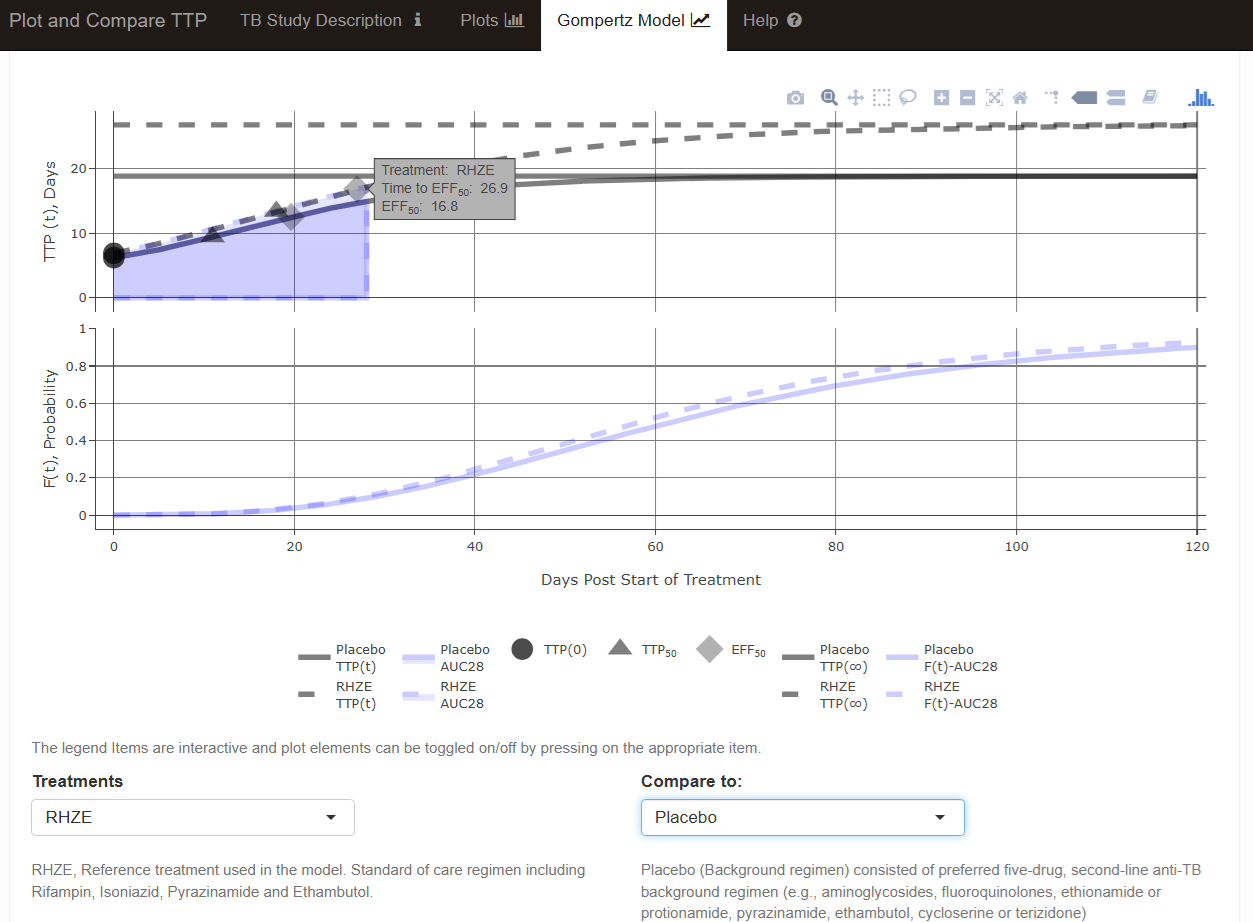
source("gompertz_helpers.R")
RHZEPRED<- makegompertzModelCurve(tvOffset=4.33843,tvAlpha=22.4067,
tvBeta=2.22748,tvGamma=0.049461)
RHZEPRED1 <- RHZEPRED[1,]
timetoeff50expression <- bquote(
"Time to"~Eff[50]==log~bgroup("(",
log~bgroup("(",bgroup("(",frac(1, 2)-frac(Offset,2%*%alpha),")")^-frac(1,beta),
")")^-frac(1,gamma),
")")
)
timetottp50expression <- bquote(
"Time to"~TTP[50]==log~bgroup("(",
log~bgroup("(",
bgroup("(",
frac(1, 2)%*%bgroup("(",1+exp(-beta),")"),
")")^-frac(1,beta),
")")^-frac(1,gamma),
")")
)
gompertzplot<- ggplot(RHZEPRED,aes(Time,TTPPRED) )+
geom_hline(aes(yintercept=Offset+Alpha)) +
geom_hline(aes(yintercept=Offset+Alpha*exp(-Beta))) +
geom_segment(data=RHZEPRED1,aes(x=TIMETTPMAX50,xend=TIMETTPMAX50,
y= TTPMAX50,yend=-Inf,
linetype=TRT),
arrow = arrow(length = unit(0.03, "npc")),
show.legend = FALSE)+
geom_segment(data=RHZEPRED1,aes(x=TIMEEFFMAX50,xend=TIMEEFFMAX50,
y= EEFFMAX50,yend=-Inf,
linetype=TRT),
arrow = arrow(length = unit(0.03, "npc")),
show.legend = FALSE)+
geom_segment(data=RHZEPRED1,aes(x=TIMETTPMAX50,xend=-Inf,
y= TTPMAX50,yend=TTPMAX50,
linetype=TRT),
arrow = arrow(length = unit(0.03, "npc")),
show.legend = FALSE)+
geom_segment(data=RHZEPRED1,aes(x= TIMEEFFMAX50, y= EEFFMAX50,linetype=TRT,
xend=-Inf,yend=EEFFMAX50),
arrow = arrow(length = unit(0.03, "npc")),
show.legend = FALSE)+
geom_text(data=RHZEPRED1,x=Inf,aes(y=(tvOffset+tvAlpha)*1.02),
label="TTP(infinity)==Offset + alpha", parse = TRUE ,
hjust=1,size=3)+
geom_text(data=RHZEPRED1,x=Inf,
aes(y=(Offset+Alpha*exp(-Beta))*0.9),
label="TTP(0)==Offset + alpha%*%exp(-beta)" ,
parse = TRUE,hjust=1,size=3)+
geom_text(data=RHZEPRED1,x=Inf,aes(y=(EEFFMAX50) ),
label="Delta==alpha-alpha%*%exp(-beta)" , parse = TRUE,
hjust=1,vjust=0,size=3)+
geom_text(data=RHZEPRED1,
x=0,aes(y=TTPMAX50+0.5,
label="frac(Offset+alpha,2)"),
parse = TRUE,hjust=0,size=3)+
geom_text(data=RHZEPRED1,
x=-5,aes(y=EEFFMAX50+1.2),
label="Offset+frac(alpha,2)%*%(1+exp(-beta) )" ,
parse = TRUE,hjust=0,size=3)+
geom_line(linewidth = 1.5,alpha=0.4)+ ###
geom_point(data=RHZEPRED1,
aes(x=TIMETTPMAX50,y= TTPMAX50),
shape=24,fill="darkgray" ,size=4,alpha=0.4)+
geom_point(data=RHZEPRED1,
aes(x=TIMEEFFMAX50,y= EEFFMAX50),
shape=23,fill="darkgray",size=4 ,alpha=0.8)+
scale_fill_manual(values=c("darkgray","darkgray"),
labels=c(expression(TTP[50]),expression(Eff[50])))+
labs(y="TTP (t), Days",x="Days Post Start of Treatment",shape="",fill="",
linetype="Parameter fold change")+
scale_x_continuous(breaks=c(0,30,60,90,120))+
coord_cartesian(xlim=c(0,120))+
theme_bw(base_size = 12)+
theme(legend.key.width = unit(1, "cm"),legend.position="top",
legend.box = "horizontal",axis.title=element_text(size=12),
axis.text=element_text(size=12))+
coord_cartesian(ylim=c(5,28))+
scale_y_continuous(breaks=c(6.891151,13.39846, 16.84404, 26.79693),
labels=c(
expression(TTP(0)),
expression(TTP[50]),
expression(Eff[50]),
expression(TTP(infinity))
))+
geom_text(data=RHZEPRED1,
aes(x=TIMEEFFMAX50+0.5,y=EEFFMAX50-1),
label= deparse(timetoeff50expression, width.cutoff = 180),
parse = TRUE,hjust=0,size=3) +
geom_text(data=RHZEPRED1,
aes(x=TIMETTPMAX50+0.5,y=TTPMAX50-2.5),
label= deparse(timetottp50expression, width.cutoff = 180),
parse = TRUE,hjust=0,size=3)