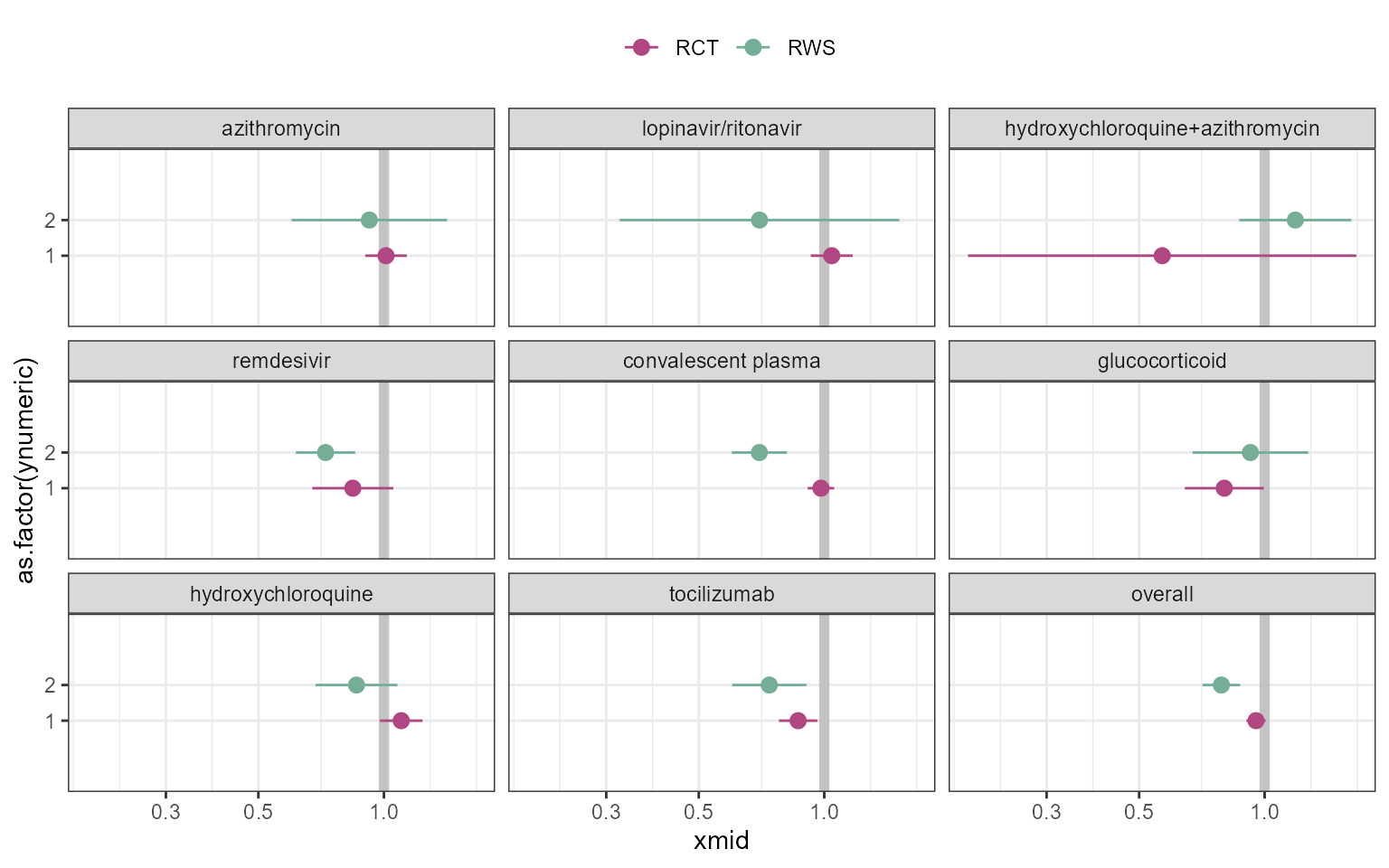
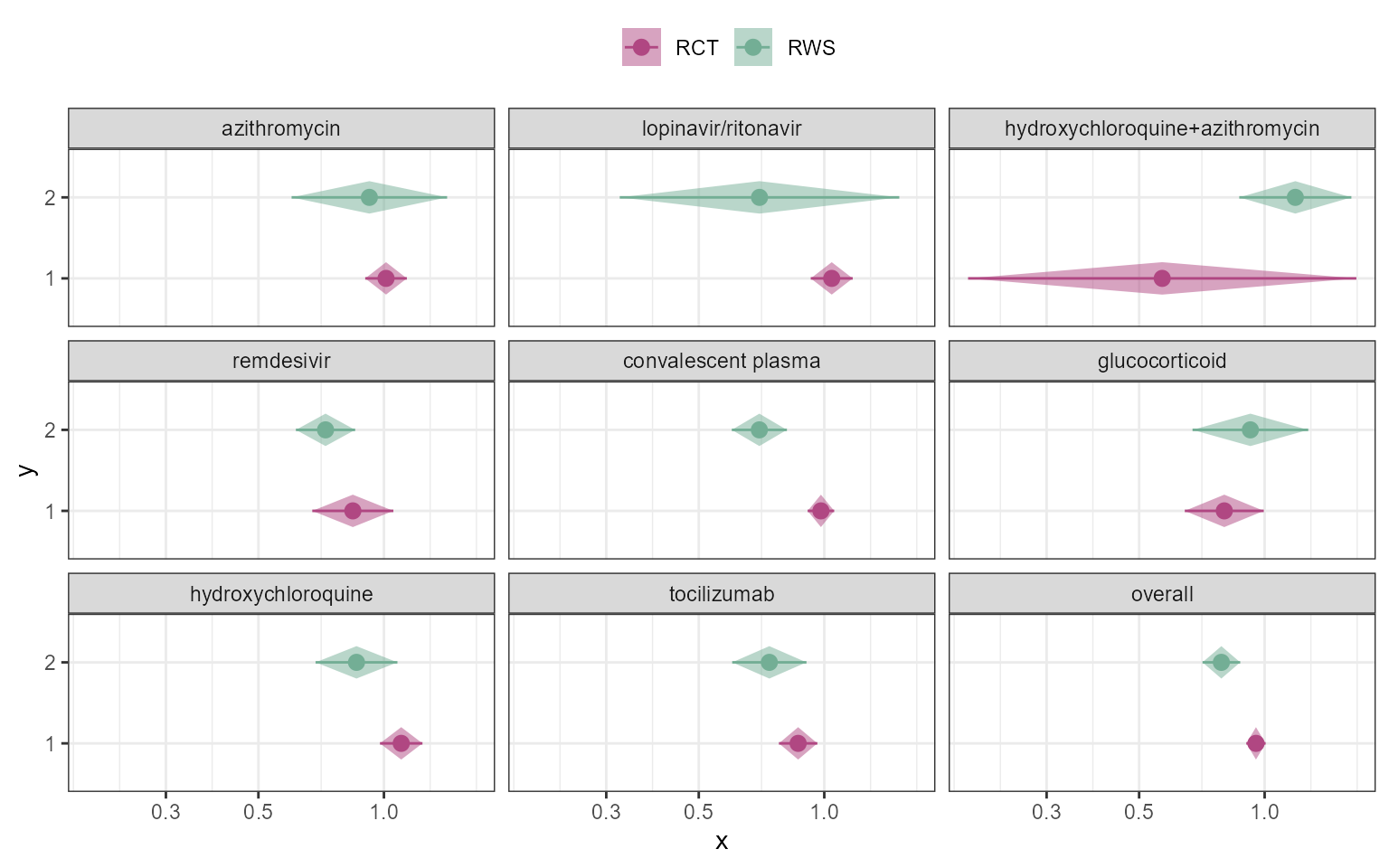
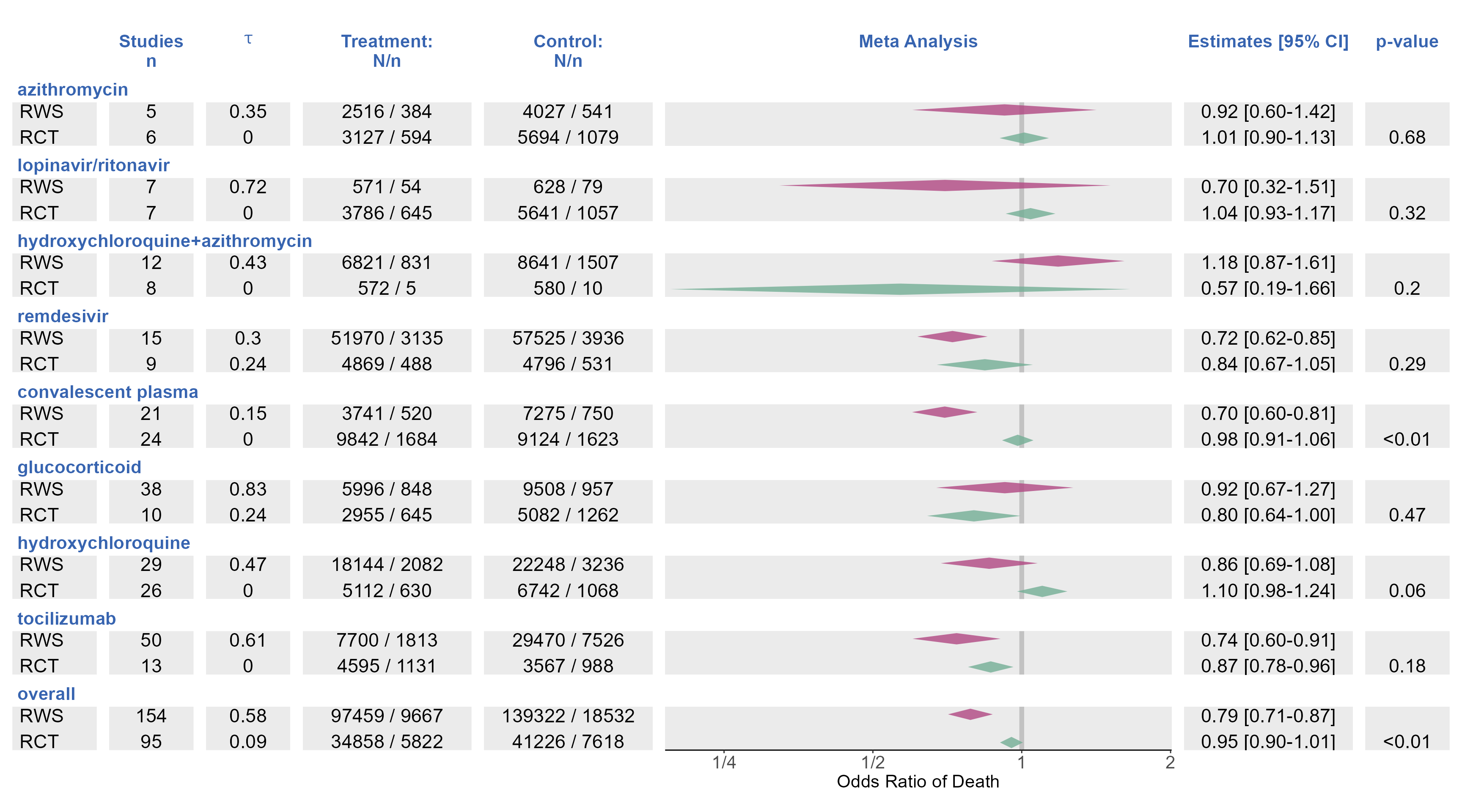
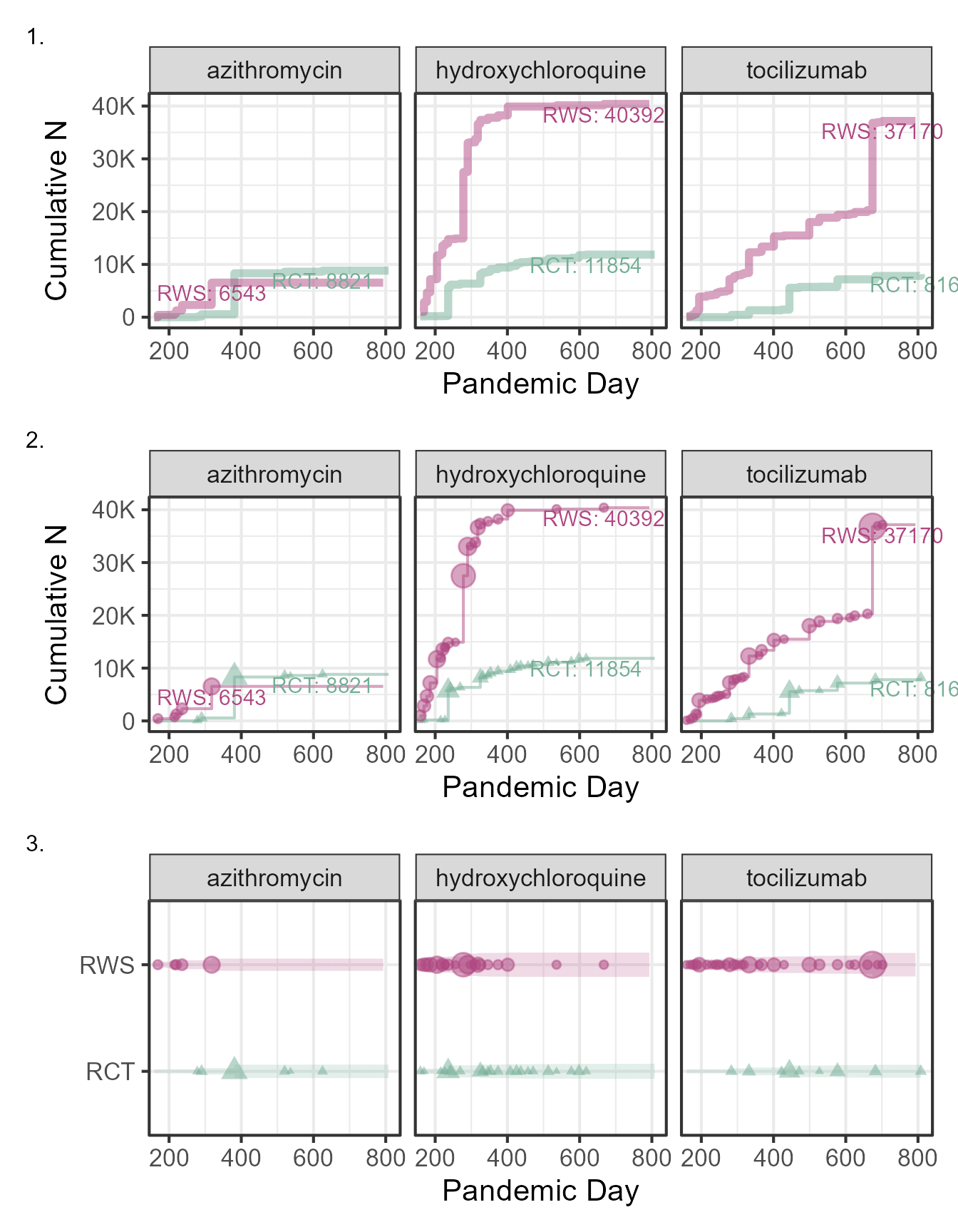
p_trt_name <- ggplot(data=figure2data)+
geom_text(aes(label=rct,y=ynumeric),x=0,hjust=0,vjust="inward",size=6, parse = TRUE)+
facet_wrap(facet~.,strip.position = "top",ncol=1,scales = "free_y")+
labs(title= expression(bold(paste(" "))))+
scale_x_continuous(expand = c(0.1,0,0.1,0),
limits=c(0,0.6),
breaks = NULL)+
theme1
p_studies <- ggplot(data=figure2data,)+
geom_text(aes(label=N,y=ynumeric),
x=0.5,hjust=0.5,size=6,vjust="inward")+
facet_wrap(facet~.,strip.position = "top",ncol=1,scales = "free_y")+
labs(title=" ",
subtitle= "Studies\nn")+
scale_x_continuous(expand = c(0,0,0,0),limits=c(0,1),breaks = NULL)+
theme2
p_trt_N <- ggplot(data=figure2data[,],)+
geom_text(aes(label=paste(Nendpoint,Nevent,sep=" / "),y=ynumeric),
x=0.5,hjust=0.5,size=6,vjust="inward")+
facet_wrap(facet~.,strip.position = "top",ncol=1,scales = "free_y")+
labs(title=" ",
subtitle= "Treatment:\nN/n")+
scale_x_continuous(expand = c(0,0,0,0),limits=c(0,1),breaks = NULL)+
theme2
p_ctl_N <- ggplot(data=figure2data,)+
geom_text(aes(label=paste(Ncontrol ,Neventcontrol,sep=" / "),y=ynumeric),
x=0.5,hjust=0.5,size=6,vjust="inward")+
facet_wrap(facet~.,strip.position = "top",ncol=1,scales = "free_y")+
labs(title=" ",
subtitle= "Control:\nN/n")+
scale_x_continuous(expand = c(0,0,0,0),limits=c(0,1),breaks = NULL)+
theme2
p_tau <- ggplot(data=figure2data,)+
geom_text(aes(label=paste( round(tau,2)),y=ynumeric),
x=0.5,hjust=0.5,size=6,vjust="inward")+
facet_wrap(facet~.,strip.position = "top",ncol=1,scales = "free_y")+
labs(title=" ",
subtitle= expression(bold(tau)))+
scale_x_continuous(expand = c(0,0,0,0),limits=c(0,1),breaks = NULL)+
theme2
p_diamonds <- plot_forest_diamond(data = figure2data,
logx=TRUE, greektau = TRUE,
xlimits = c(0.19,2.01),
xbreaks = c(1/8,1/4,1/2,1,2),
xlabels = c("1/8","1/4","1/2","1","2"),
expandleft=0,
expandright=0,
stripx.text.color="transparent")+
labs(title=" ",subtitle= "Meta Analysis")+
theme2
p_95 <- ggplot(data=figure2data,)+
geom_text(aes(label=LABEL,y=ynumeric),
x=0.5,hjust=0.5,size=6,vjust="inward")+
facet_wrap(facet~.,strip.position = "top",ncol=1,scales = "free_y")+
labs(title=" ",
subtitle = expression(bold(paste("Estimates [95% CI]"))))+
scale_x_continuous(expand = c(0,0,0,0),limits=c(0,1),breaks = NULL)+
theme2
p_pvalue <- ggplot(data=figure2data,)+
geom_text(aes(label=ifelse(p<0.01,"<0.01",p),y=ynumeric),
x=0.5,hjust=0.5,size=6,vjust="inward")+
facet_wrap(facet~.,strip.position = "top",ncol=1,scales = "free_y")+
labs(title=" ",
subtitle= expression(bold(paste("p-value"))))+
scale_x_continuous(expand = c(0,0,0,0),limits=c(0,1),breaks=NULL)+
theme2
#ggsave("forestplot_redo.png",foresplot,width =20 ,height = 10.4, dpi = 300)
(p_trt_name | p_studies |p_tau |p_trt_N | p_ctl_N |p_diamonds| p_95|p_pvalue)+
plot_layout(widths = c(0.1,0.1,0.1,0.2,0.2,0.6,0.2,0.1))